-Search query
-Search result
Showing 1 - 50 of 101 items for (author: abraham & j)

EMDB-15786: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15787: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M
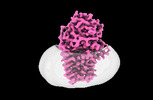
EMDB-15788: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15789: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M
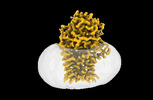
EMDB-15790: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15791: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0k: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0l: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0m: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0n: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0o: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0p: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-29044: 
Structure of Zanidatamab bound to HER2
Method: single particle / : Worrall LJ, Atkinson CE, Sanches M, Dixit S, Strynadka NCJ

EMDB-13738: 
Horse spleen apoferritin
Method: single particle / : Koning RI, Renault L

EMDB-25673: 
Prepore structure of pore-forming toxin Epx1
Method: single particle / : Xiong XZ, Yang P

PDB-7t4e: 
Prepore structure of pore-forming toxin Epx1
Method: single particle / : Xiong XZ, Yang P, Dong M, Abraham J

EMDB-13654: 
DNA polymerase from M. tuberculosis
Method: single particle / : Borsellini A, Lamers MH

PDB-7pu7: 
DNA polymerase from M. tuberculosis
Method: single particle / : Borsellini A, Lamers MH

EMDB-13102: 
human LonP1, R-state, incubated in AMPPCP
Method: single particle / : Abrahams JP, Mohammed I, Schmitz KA, Schenck N, Maier T

PDB-7oxo: 
human LonP1, R-state, incubated in AMPPCP
Method: single particle / : Abrahams JP, Mohammed I, Schmitz KA, Schenck N, Maier T

EMDB-25209: 
Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab
Method: single particle / : Pan J, Abraham J, Yang P, Shankar S

EMDB-25210: 
Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region)
Method: single particle / : Pan J, Abraham J, Shankar S

PDB-7sn2: 
Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab
Method: single particle / : Pan J, Abraham J, Yang P, Shankar S

PDB-7sn3: 
Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region)
Method: single particle / : Pan J, Abraham J, Shankar S

EMDB-24896: 
CryoEM structure of Gq-coupled MRGPRX2 with peptide agonist Cortistatin-14
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

EMDB-24897: 
CryoEM structure of Gi-coupled MRGPRX2 with peptide agonist Cortistatin-14
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

EMDB-24898: 
CryoEM structure of Gq-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

EMDB-24899: 
CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

EMDB-24900: 
CryoEM structure of Gq-coupled MRGPRX4 with small molecule agonist MS47134
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

PDB-7s8l: 
CryoEM structure of Gq-coupled MRGPRX2 with peptide agonist Cortistatin-14
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

PDB-7s8m: 
CryoEM structure of Gi-coupled MRGPRX2 with peptide agonist Cortistatin-14
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

PDB-7s8n: 
CryoEM structure of Gq-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

PDB-7s8o: 
CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

PDB-7s8p: 
CryoEM structure of Gq-coupled MRGPRX4 with small molecule agonist MS47134
Method: single particle / : Cao C, Fay JF, Gumpper RH, Roth BL

EMDB-23373: 
Negative stain EM map of RM20C Fab in complex with edc BG505 SOSIP.664
Method: single particle / : Turner HL, Ward AB, Cottrell CA

EMDB-12313: 
P2c-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12315: 
R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12316: 
D1-state of wild type human mitochondrial LONP1 protease
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12317: 
Human mitochondrial Lon protease homolog, D2-state
Method: single particle / : Mohammed I, Abrahams JP, Schmitz KA, Maier T, Schenck N

PDB-7ngf: 
P2c-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

PDB-7ngl: 
R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

PDB-7ngp: 
D1-state of wild type human mitochondrial LONP1 protease
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

PDB-7ngq: 
Human mitochondrial Lon protease homolog, D2-state
Method: single particle / : Mohammed I, Abrahams JP, Schmitz KA, Maier T, Schenck N

EMDB-12696: 
Amyloid beta oligomer displayed on the alpha hemolysin scaffold
Method: single particle / : Wu J, Blum TB, Farrell DP, DiMaio F, Abrahams JP, Luo J

PDB-7o1q: 
Amyloid beta oligomer displayed on the alpha hemolysin scaffold
Method: single particle / : Wu J, Blum TB, Farrell DP, DiMaio F, Abrahams JP, Luo J

EMDB-12312: 
P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

PDB-7ngc: 
P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12306: 
P1a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and ATPgS
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12307: 
P1b-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP

EMDB-12308: 
P1c-state of wild type human mitochondrial LONP1 protease with bound substrate protein in presence of ATP/ADP mix
Method: single particle / : Mohammed I, Schmitz KA, Schenck N, Maier T, Abrahams JP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
